Fungi is a kingdom
from one of the five eminent kingdoms of life
easy to access and a open repository of fungi kingdom
Every molecule in a cell has its special characteristics, including the individual proteins that comprise the proteome of an organism. Proteomes comprise all of the translated products of nucleotide sequences encoded in the messenger RNA (mRNA). The total mRNA of an organism encodes wide arrays of proteins that vary in cellular function and homeostasis. These proteins have diverse molecular masses and isoelectric points (pIs). Post-translational modifications can alter a protein's function and contribute to the ability to target the location of a protein to a specific subcellular compartment. The shape, size, solubility, and pI of a protein determine its ability to move across different cellular compartments and also determine its function. Fungi cells contain many proteins with different molecular masses and pI. The pI indicates the pH at which the net charge of a protein is zero. The dissociation constant (pKa) of a polypeptide is determined by the presence of seven charged amino acids; arginine, aspartate, cysteine, glutamate, histidine, tyrosine, and lysine.
The N-terminal NH2- and C-terminal COOH-group of a protein also influence the charge of a polypeptide. Post-translational modifications, protein-protein interactions, dipole interactions, and other biochemical factors also affect the pI of a protein. Molecular mass and pI are used to determine the position of a protein sequence in a proteome map and provide helpful information to bioinformatics and genome scientists seeking to understand the molecular basis of subcellular localization and function. Several attempts have been made to create a database of experimentally validated proteins. However, it isn't easy to experimentally validate each protein's pI and molecular mass in a proteome.
Fungi Database is formed in main 3 sections
An exclusive and aggregate summary statistics about maximum, minimum and average values of species proteomes, proteins, sequences, isoelectric point (pI) including its pI types and molecular weights.
All 685 species with core attributes (i.e. species name, total number of proteins, total number of basic pI proteins, total number of neutral proteins, total number of acidic proteins etc.) of fungi kingdom.
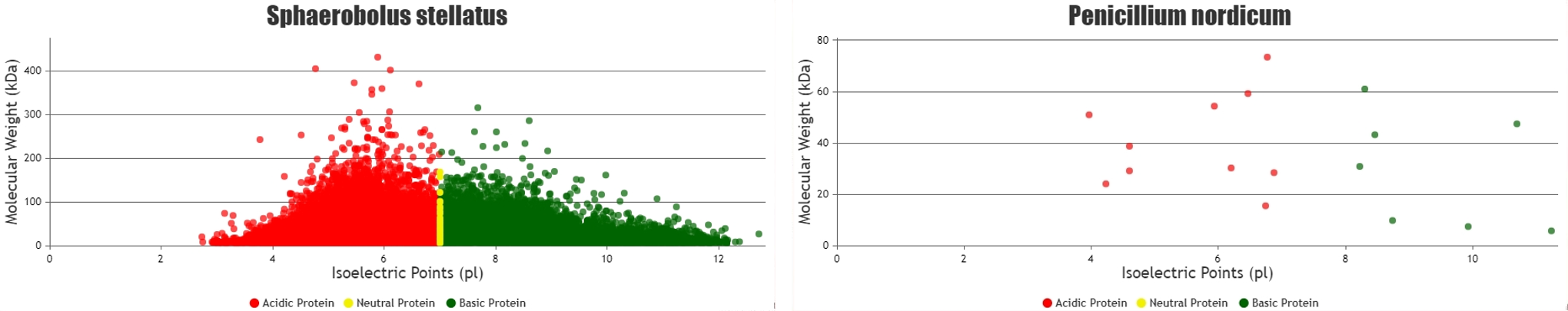
Two species (sphaerobolus stellatus & trichoderma asperellum) scatter plots with maximum and minimum number of proteins respectively.
This is the main and core section of fungi database that contains each species detailed proteomic information. It contains each species detailed proteomic information i.e. accession numbers, sequences, name of proteins, molecular weights, isoelectric points (pI).